Brief Description
-
Why was this study conducted?
Despite multiple studies associating various clinical characteristics with developing GDM, many of them are limited in their retrospective nature or number of variables modeled. We aimed to prospectively recruit a cohort with a robust collection of survey, biomarker, wearable, and other data to be able to perform a more comprehensive GDM prediction model. -
What are the key findings?
Through stepwise logistic regression: prior history of GDM, current diagnosis of chronic hypertension, higher BMI, and higher insomnia scores were found to be independently associated with GDM. -
What does this study add to what is already known?
The study data collected will be paired with multi-omics data to develop clinically useful GDM prediction tools and will be available for collaborative secondary analyses.
Introduction
Gestational diabetes mellitus (GDM) is a transient form of diabetes that develops during pregnancy, affecting up to 15% of pregnancies (Modzelewski et al. 2022; “ACOG Practice Bulletin No. 190: Gestational Diabetes Mellitus” 2018). GDM raises the risk of adverse perinatal outcomes, and at least 20% of women with GDM will develop type 2 diabetes mellitus (T2DM) within 10 years, with a lifetime risk of up to 70% (“ACOG Practice Bulletin No. 190: Gestational Diabetes Mellitus” 2018; Z. Li, Cheng, Wang, et al. 2020). Risk factors for GDM include elevated pre-pregnancy body-mass index (BMI), weight gain during pregnancy, greater maternal age, as well as higher plasma triglyceride and lower HDL-C levels (Noctor and Dunne 2015). Children born to mothers with GDM also have higher birth weight, carry a greater risk of childhood obesity, and are more likely to develop T2DM in adult life (Nijs and Benhalima 2020).
Despite multiple studies associating various clinical characteristics with developing GDM, many of them are limited in their retrospective nature or limited number of variables that are based only on routine clinical measurements available in the electronic health record (Artzi, Shilo, Hadar, et al. 2020). Inclusion of not only clinical characteristics, but also objectively measured behavioral characteristics such as physical activity, could improve the predictive accuracy and serve as a potential basis for preventive interventions. Additionally, the use of genetics and the tools of machine learning in predictive model development have also been able to refine and improve predictive models for other adverse pregnancy outcomes (Marić, Tsur, Aghaeepour, et al. 2020; Schmidt, Rieger, Neznansky, et al. 2022).
The objective of this study was to prospectively recruit a pregnancy cohort early in gestation to develop a comprehensive set of associated features with the development of GDM. These features, including clinical, sociodemographic, behavioral, biochemical, and genomic, could then be utilized in a machine learning predictive modeling study to not only predict GDM with more precision, but also provide therapeutic targets for pre-pregnancy and early pregnancy interventions to decrease the impact of GDM on health during and after pregnancy.
Materials and Methods
Study participants
This study was a prospective observational cohort study of pregnant individuals, titled the Hoosier Moms Cohort (HMC). The HMC included individuals with a singleton gestation, with a gestational age of less than 20 weeks confirmed by the American College of Obstetrics and Gynecology (ACOG) ultrasound dating guidelines, and who were at least 18 years old at the time of consent. Individuals with any type of pre-gestational diagnosis, HbA1c at screening of ≥6.5 (which was performed at study screening to exclude individuals with undiagnosed pregestational diabetes), or abnormal 3-hour oral glucose tolerance test before 20 weeks of gestation were excluded. Other exclusion criteria included pre-pregnancy chronic systemic steroid use, planned pregnancy termination, inability to provide informed consent in English or Spanish or to complete longitudinal study activities, and presence of major fetal anomalies prior to enrollment. All participants provided written informed consent. The study was approved by the Indiana University Institutional Review Board.
Participants were recruited via self-referral in response to advertisements and from clinics associated with providers from the Indiana University School of Medicine in the Indianapolis area and from Deaconess Health in Evansville, Indiana. Study team members preferentially recruited pregnant individuals at high risk of developing GDM (e.g. with obesity) but recruited any eligible individual.
We initially aimed to recruit 500 pregnant participants early in their pregnancy, but due to the pandemic and funding, recruitment was stopped early after 411 participants were enrolled. No a priori sample size calculation for a pre-defined outcome was performed. We anticipated that, recruiting a higher-risk population for GDM (based on our clinical sites), we would expect about 10-16%, or 50-80 participants, to develop GDM.
Subjects were withdrawn and replaced for failure to complete visit 1 activities- surveys, biospecimens, and biometrics (all three had to be missed for administrative withdrawal) or if later ultrasound found them to be ≥ 20 weeks gestation at enrollment.
Visits
If not already performed clinically, a dating ultrasound was performed for the study for accurate pregnancy dating. Subjects completed study activities at up to eight time points over the course of two years: two visits in the prenatal period (V1[<20 weeks] and V2 [24-32 weeks]), and at Delivery (V3). We planned one visit at 4 to 16 weeks Postpartum (V4), four online/phone Interval Contacts occurring between 6 months and 18 months after delivery (M6, M12, M18 & M24), and one visit at Year 2 (V5). Due to funding constraints, V5 visits were only performed for participants who developed GDM or had been diagnosed with diabetes, pre-diabetes, or metabolic syndrome postpartum.
Study activities
Subject interviews, self-administered surveys, and biospecimen collections were performed as listed in Table 1.
Biologic samples (blood, urine, stool) were taken at visits 1-5, and surveys were administered at visits 1,2,4 and 5. Participant biometrics recorded included height, weight, heart rate, blood pressure, waist and hip measurements, and body composition. These were collected by trained study team members or clinical staff using standard clinical instruments. Additionally, placental samples and cord blood were collected at delivery. Blood was collected for DNA extraction as well as processed in standard fashion and aliquoted for plasma and serum. All specimens were stored at -80° Celsius until analyses. A blood sample was also sent to a research lab at baseline for measures of hemoglobin A1c (HbA1c), a lipid panel, as well as a urine specimen for a baseline protein/creatinine ratio. Infant cord blood (or alternate buccal swabs if cord blood was unable to be obtained at delivery) was also obtained for DNA extraction. DNA extraction and genotyping procedures are described below.
All participants were provided with a Garmin VivoFit 4 activity tracker and instructed on its use. They were encouraged to wear it as much as possible and to synchronize with the server routinely. Participants were called 1 week before any planned study visits to encourage them to wear the device the week before the visit to have contemporaneous data with the visit. The device captured physical activity data and sleep data (when worn at night). Access to the raw data in the Garmin data warehouse was obtained. Physical activity data were converted to daily and weekly metabolic equivalents (METs) for analysis. Measures of sleep and vital signs (heart rate, blood pressure, pulse oxygenation) during these times were also available for analysis.
Foodprint app: A photo-based food diary and visual summary system was developed by a group led by one of the co-authors (CC) (Chung, Wang, Schroeder, et al. 2019). The app was adapted from the designs of other photo-based diaries to focus on helping people communicate their healthy eating goals and progress with health experts. This consisted of three tools: (1) a mobile app supporting in-the-moment, low-burden food capture, (2) a web app presenting relationships between food and health goals, and (3) a pre-visit note that asked participants to summarize their data for the visit. Participants downloaded the app with the study team, were assigned a study ID for their data, given instructions on synching, and were asked to use the app as much as possible and tolerable. Additionally, they were contacted before visits and encouraged to track their food carefully during the week before. More details about the app can be found in the development paper (Chung, Wang, Schroeder, et al. 2019).
Surveys: As per Table 1, participants were asked questions about medical history, behaviors, pregnancy history, as well as validated surveys. Diet was assessed by the Automated Self-Administered 24-hour (ASA24)® Dietary Assessment Tool (Subar, Kirkpatrick, Mittl, et al. 2012). The Three Factor Eating Questionnaire (TFEQ) (Stunkard and Messick 1985) and Modifiable Activity Questionnaire (MAQ) (Gabriel et al. 2010) were also administered. Depression symptoms were measured by the Edinburgh Postnatal Depression Scale (EPDS) (Cox et al. 1996). The Perceived Stress Scale (PSS) (Cohen, Kamarck, and Mermelstein 1983), Pregnancy Experiences Scale (PES, brief version) (DiPietro, Christensen, and Costigan 2008), Women’s Health Initiative Insomnia Rating Scale (WHIIRS) (Levine et al. 2003), Berlin Questionnaire for Sleep Apnea (Netzer et al. 1999), London Measure of Unplanned Pregnancy (Barrett, Smith, and Wellings 2004), and Adverse Childhood Experience (ACE) Questionnaire (Meinck et al. 2017) were all administered per Table 1. All surveys were selected as they were validated instruments that had been used in other pregnancy cohorts.
After delivery, chart abstractions for outcomes were conducted by trained and certified abstractors at least 30 days postpartum. Pregnancy and newborn outcomes were abstracted, including clinical laboratory values, pregnancy-related conditions (including GDM), gestational age at birth, newborn weight and body measurements, and newborn outcomes. GDM was diagnosed in all individuals using 2-step screening with the Carpenter-Coustan criteria at the transition to the 3rd trimester (“ACOG Practice Bulletin No. 190: Gestational Diabetes Mellitus” 2018).
Genetic studies
DNA extraction and genotyping: We genotyped participants who had contributed whole blood samples during their first visit (n = 393). DNA extractions were carried out on a QIAsymphony instrument (from Qiagen; extraction kit DSP DNA Midi Kit #937355, protocol Blood_1000_V7_DSP) at the Center for Genomics and Bioinformatics (Indiana University, Bloomington), and genotyping was completed at the Van Andel Institute (Grand Rapids, MI, USA). Genotyping was carried out using the Infinium™ Global Diversity Array-8 v1.0 with 1,825,277 markers (Illumina, Miami, USA). Raw intensity data (.idat files) were inspected and filtered with GenomeStudio v2.4 (Illumina). We carried out initial quality control using standard technical filters: cluster separation < 0.3, normalized R-value mean < 0.2 for all genotypes, and 10th percentile of the GenCall scores < 0.3). Genotype calls for the 1,768,794 loci that passed initial quality control (97% of all markers in the array) were made with Beeline autoconvert (Ilumina). These files were then converted to Variant Call Format (VCF) using the gtc2vcf plugin from bcftools (https://github.com/freeseek/gtc2vcf). The vcf file was processed to only retain single nucleotide variants with genotyping rate > 95%, minor allele frequency > 0.01, and that were under Hardy-Weinberg equilibrium (i.e. HWE P > 5×10−2). After imposing these filters, 845,841 SNP markers remained. The vcf file was also examined to exclude any individuals with a call rate less than 98 % (note that all 393 individuals genotyped passed this filter). The filtered vcf was phased with EAGLE and imputed via the TOPMED Imputation Server version R2.
Association Testing and Polygenic Risk Scores (PRS): The sample size of our cohort would generally not be large enough to conduct a genome-wide search for association with GDM, so we used the imputed genetic data in two ways: 1) to validate loci previously associated with diabetes, and 2) to evaluate the performance of reported diabetes PRS (Powe, Nodzenski, Talbot, et al. 2018; Polfus, Darst, Highland, et al. 2021).
We compiled a list of 673 genetic markers from three publicly available sources: (1) 21 reported GDM associations in the NHGRI-EBI GWAS catalog (trait ID EFO_0004593); (2) 582 markers associated with T2D or GDM and included in the PRS derived by Polfus et al. (Polfus, Darst, Highland, et al. 2021); (3) 79 markers not included in the previous two but included in the PRS derived by Powe et al (Powe, Nodzenski, Talbot, et al. 2018). We used bcftools to extract all markers of interest that were present in our imputed data set (20, 582, and 71 markers from each source, respectively) (Danecek, Bonfield, Liddle, et al. 2021). We tested for genetic association to GDM using PLINKv1.9 with 16 individuals excluded due to missing phenotype data (n= 40 cases and 344 controls) (Purcell, Neale, Todd-Brown, et al. 2007). We fit a logistic regression model and included the first principal components of genetic variation as covariates to control for population stratification. Principal components were derived using PLINK on a pruned set (i.e. taking windows of independent markers genome-wide; linkage disequilibrium r2 < 0.5).
We then evaluated the performance of available PRS for T2D on our cohort, an approach successfully used by Powe et. al (Powe, Nodzenski, Talbot, et al. 2018). PRS was calculated for the cohort via PLINK v1.9 (6) with the effect sizes of the 582 markers provided by Polfus et al (Polfus, Darst, Highland, et al. 2021). Individuals with missing phenotype data were excluded from the analysis.
Since the available PRS was derived from a cohort of individuals with European ancestry, we only evaluated the score for individuals with high similarity to the EUR superpopulation of the 1000 Genomes Consortium (Auton, Brooks, Durbin, et al. 2015). We inferred genetic similarity using SNPweights v.2.1, and set the threshold of probability of assignment to a cluster at >51%.
Overall Statistical Analysis
We performed descriptive analyses of the cohort and compared the characteristics of those who did and did not develop GDM. Continuous variables were analyzed using either two-sample t-test or Wilcoxon Rank Sum test. Categorical variables were compared using chi-square test or Fishers Exact test. Multiple logistic regression analysis was utilized to find independent associations of characteristics with GDM development. A stepwise logistic regression was performed, starting with all variables that had a univariable association p-value of ≤ 0.10.
Results
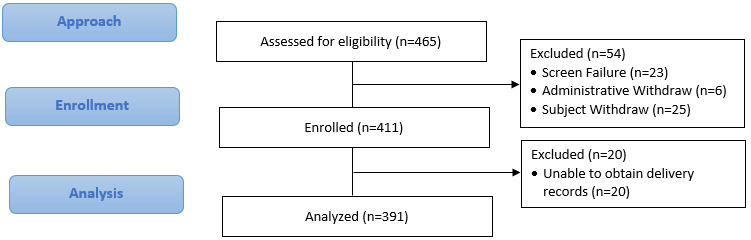
A total of 411 participants were recruited into the HMC (Figure 1). Of those participants, complete outcome data were available for 391 (95.1%). Outcome data were missing for some participants who delivered outside of the study area, and we were unable to obtain medical records for the primary outcome or who were lost to follow-up before delivery. Characteristics of the cohort are given in Table 2. Patients were on average 30 years of age, had a mean BMI of 28, and 17% were of Hispanic ethnicity. Study participants mostly racially identified as White (71%), followed by Black (16%). V1 questionnaires showed that 63% of participants were nulliparous. Additionally, 54% reported a family history of diabetes, with 4% of patients reporting a personal prior history of GDM.
A total of 39 participants (10.0%) developed GDM. Compared to those that did not, participants who developed GDM had a significantly higher baseline BMI (31.6 vs 27.2, p=0.003), HbA1c (5.24 vs 5.07, p<0.001), triglycerides (156.8 vs 134.2, p=0.022), and blood glucose (85.90 vs 79.96, p=0.025) at the initial visit (V1). They also had a lower probability of being nulliparous prior to this pregnancy (15.38% vs 33.61%, p=0.02), and had a higher chance of having a prior history of gestational diabetes (28.21% vs 1.96%, p<0.0001), and current chronic hypertension (12.82% vs 1.9%, p=0.0034). Additionally, they scored higher on the WHIIRS questionnaire (9.62 vs 7.80, p=0.028).
During the study, the GDM group had a significantly higher rate of developing a hypertensive disorder of pregnancy (43.59% vs 26.05%, p=0.02), while also having an earlier EGA at delivery (38.52 vs 39.59 weeks, p=0.01).
We found a significant association with GDM in 3 out of 673 previously reported genetic markers (P < 0.01; Table 3). Individuals with high polygenic risk scores were not more likely to have a GDM diagnosis. The rates of GDM in both the lowest PRS risk quartile and highest risk quartile were similar, 13.6% and 13.8%, respectively (Figure 2; n=261 individuals classified as EUR). Controlling for multiple variables above, the stepwise logistic regression found that prior history of GDM, current diagnosis of hypertension, insomnia, and BMI respectively were the most impactful variables in our best model fit (Table 4).
Structured Discussion
a. Principal Findings
The clinical prediction and prevention of GDM are important in managing both pregnancy outcomes and the potential lifelong sequalae that come with the diagnosis. There are a host of preestablished risk factors for GDM, notably BMI and prior history of GDM, that help clinicians guide counseling and potential early screening. Our study included multiple other characteristics, biomarkers, genetics, and lifestyle characteristics to improve upon currently known associations. Higher pre-pregnancy BMI, history of GDM in a prior pregnancy, being primigravida, and chronic HTN are well known risk factors for developing GDM in a subsequent pregnancy (Y. Zhang, Xiao, Zhang, et al. 2021; Muche, Olayemi, and Gete 2019). Our results also reaffirm the increased incidence of preterm delivery and the development of hypertensive disorders of pregnancy in pregnancies complicated by GDM (Ye et al. 2022; Yogev, Xenakis, and Langer 2004). Additional risk factors found statistically relevant in our study were higher early pregnancy HbA1c, triglycerides, and blood glucose values in the group that subsequently developed GDM.
b. Results in the Context of What is Known
A risk prediction model for GDM was recently developed using a nomogram, and subsequently identified common risk factors, such as age, BMI, family history of GDM, and fasting blood glucose, in addition to others such as hemoglobin and serum ferritin that were not included in our analysis (R. Li et al. 2023). This model was proven successful with the area under the curve in the training group was 0.920, and that of the validation group was 0.753 (R. Li et al. 2023). We plan to utilize the additional data collected (including multiple -omics work currently underway) in the Hoosier Moms Cohort and machine learning techniques to develop a predictive model that includes characteristics not ascertained in other studies.
Another maternal characteristic that was associated with the development of GDM independently was the WHIIRS score at the first visit. Insomnia in pregnancy, leading to short sleep duration, has been associated with GDM (Facco, Chan, and Patel 2022). Insomnia is also associated with increased perinatal anxiety and depression in patients with and without GDM (Facco, Chan, and Patel 2022; Aydin and Dogru 2022). A systematic review found that anxiety and depression independently increase the risk of developing GDM (OuYang et al. 2021). Understanding the intricate interplay of these predictive factors and how to address them both before and during pregnancy will be important reduce the incidence of GDM.
The variables identified through logistic regression to have the most impact on developing GDM are similar to those identified in some other studies. A cohort study evaluated whether recurrent GDM and new GDM diagnoses shared similar risk factors (L. Zhang et al. 2022). They found that the risk of GDM in subsequent pregnancies increased threefold with a previous GDM diagnosis (L. Zhang et al. 2022). These findings are also reflected in another that demonstrated an association between a previous diagnosis of GDM and the development of GDM (p= 0.0001) (Kouhkan, Najafi, Malek, et al., n.d.). In our HMC group, the independent odds for GDM in participants with a prior history was increased nearly 15-fold when controlling for all other characteristics. In our cohort, a diagnosis of chronic hypertension was also independently associated with GDM, similar to a different cross-sectional study (Aburezq et al. 2020). Elevated BMI and Insomnia have also been associated with GDM (Kouhkan, Najafi, Malek, et al., n.d.; Myoga, Tsuji, Tanaka, et al. 2019).
c. Clinical Implications
Various efforts have been made to develop an innovative prediction model for GDM. Researchers in Mexico collected data from 1709 pregnant patients and selected the best predictive variables through a machine-learning-driven method, such as age, BMI, parity, and capillary blood glucose at the first visit (Gallardo-Rincón, Ríos-Blancas, Ortega-Montiel, et al. 2023). An artificial neural network approach was used to build a model achieving high levels of accuracy and sensitivity for identifying women at a high risk of developing GDM (Gallardo-Rincón, Ríos-Blancas, Ortega-Montiel, et al. 2023). This model is touted to be simple and easy to implement even in low-resource settings, however, the authors were skeptical as to the generalization of the model (Gallardo-Rincón, Ríos-Blancas, Ortega-Montiel, et al. 2023). Validating any model in a more diverse population will be important before widespread clinical implementation.
Our study included a genetic association analysis that found several variants positively and negatively associated with GDM. A recent GWAS of 5485 patients with GDM and 347,856 without GDM found 5 variants significantly associated with GDM (MTNR1B, TCF7L2, CDKAL1, CDKN2A-CDKN2B, and HKDC1) (Pervjakova, Moen, Borges, et al. 2022). The HKDC1 gene (rs9663238) was also associated with GDM in our data set (p=0.001554). We also found markers near TCF7L2 and MTNR1B, but not those particular genes. The differences in our findings may be due to the smaller cohort size in our study. In our cohort, the type 2 Diabetes PRS score derived by Polfus et al. was not significantly associated with meaningful differences in GDM rates (Polfus, Darst, Highland, et al. 2021). This is contrary to previous results (Powe, Nodzenski, Talbot, et al. 2018; Pagel, Chu, Ramola, et al. 2022). In patients with a genetic predisposition to GDM (based on polygenic risk scores), increased physical activity has been shown to reduce the GDM risk (Pagel, Chu, Ramola, et al. 2022). Thus, understanding genetic-based risk may be important clinically and further work is planned.
d. Research Implications
These associations with GDM include some unique features such as insomnia ratings and genetic studies. Further work into comprehensive predictive models that can lead to clinical interventions to reduce the risk of developing GDM are important. These should further explore if the addition of biochemical markers or clinical measures not part of routine care significantly improve current models. Uncertainty also remains regarding optimal timing of GDM testing, something not explored in this study.
e. Strengths and Limitations
Our study has limitations. While we were successful in recruiting a relatively high-risk cohort that ended up with a 10% incidence of GDM, the number of participants was too low for some analyses and adjusting for multiple comparisons. While our cohort was all recruited from the state of Indiana, it was diverse in sociodemographic characteristics and likely representative of many populations in the United States. The sample size limited some additional analyses of the genetic findings or many subgroup analyses. We were able to confirm several previously known clinical and genetic associations but had the added strength of combining baseline demographic predictors with lab values and psychosocial instruments to create a more robust predictive model. Future results from multiple -omics assays will be combined using cutting-edge machine learning tools to create a clinically useful predictive tool and to test its performance against common clinical characteristics. The use of these additional features, even with the relatively small sample size, will contain features not found in other cohorts.
f. Conclusions
In conclusion, the Hoosier Moms Cohort identified that participants with a previous GDM diagnosis, hypertension, elevated BMI, and insomnia have significantly increased odds of developing GDM in a diverse cohort of participants. These data, along with other biomarker assays forthcoming, will be utilized to create a clinically useful predictive tool. Given the rise in GDM rates and potential lifelong consequences for the mother and infant, it is crucial to improve our ability to predict and prevent GDM.
Authorship Contributions
David Haas: Conceptualization, Methodology, Project Administration, Investigation, Funding acquisition, Data curation, Formal analysis, Supervision, Hani Faysal: Formal analysis, Writing- original, review, editing. Mitchell Grecu: Investigation, Data curation, Writing- original, review, editing. Kathleen Flannery: Methodology, Project Administration, Investigation, Data curation, Writing- review, editing. Haley Schmidt: Project Administration, Investigation, Data curation, Writing- review, editing. Maha Aamir: Investigation, Formal analysis, Data curation, Writing- original, review, editing. Rafael Guerrero: Methodology, Investigation, Formal analysis, Data curation, Writing- original, review, editing. Chia-Fang Chung: Conceptualization, Methodology, Project Administration, Investigation, Writing- review, editing. Constantine Scordalakes: Project Administration, Investigation, Writing- review, editing. Brennan Fitzpatrick: Project Administration, Investigation, Writing- review, editing. Shelley Dowden: Project Administration, Data curation, Writing- review, editing. Shannon Barnes: Project Administration, Investigation, Data curation, Writing- review, editing. David Guise: Project Administration, Data curation, Formal analysis, Writing- review, editing. Aric J Kotarski: Project Administration, Data curation, Formal analysis, Writing- review, editing. Chandan Saha: Methodology, Project Administration, Data curation, Formal analysis, Writing- review, editing. Predrag Radivojac: Conceptualization, Methodology, Project Administration, Investigation, Funding acquisition, Writing- review, editing. Christina Scifres: Conceptualization, Methodology, Investigation, Writing- review, editing. Katherine Connelly: Conceptualization, Methodology, Project Administration, Investigation, Funding acquisition, Writing- review, editing.
Conflicts of Interest
The authors declare no conflict of interests.
Funding
Funding for the Hoosier Moms Cohort was provided by the Indiana University Grand Challenges Precision Health Initiative.
Presentation
The results were presented as a poster presentation at the 2023 Annual Meeting of the Central Association of Obstetricians and Gynecologists in Nashville, TN, October 26-27, 2023.
AI
No AI was used in the production of this manuscript.


_in_diabetes_polygenic_risk_score_(prs)_quartiles.png)
_in_diabetes_polygenic_risk_score_(prs)_quartiles.png)